UNIT 5
Enzymes
(monitoring enzyme catalysed reactions)
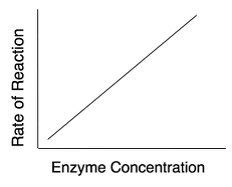
Fig1: For a give enzyme concentration, the rate of reaction increases with increase in substrate concentration until all the available active sites are occupied by the substrates.
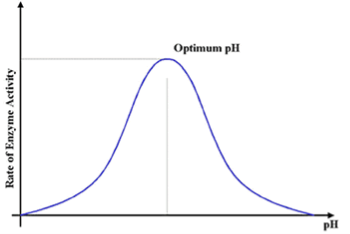
Fig 2: The rate of enzyme activity is maximum, at optimum pH.
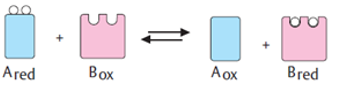
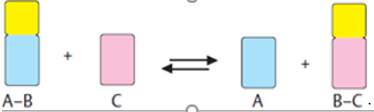
6. In general, enzymes are proteins produced by living cells, they act as catalysts in several biochemical reactions. A catalyst also affects the rate of a chemical reaction. In the presence of an Enzyme the cell can carry out complex chemical reactions at relatively low temperatures through enzyme activity. In an enzyme-catalysed reaction, the substance to be acted upon (the substrate = S) binds reversibly to the active site of the enzyme (E).The result that forms such a temporary union shows a reduction in the energy required to activate or proceed the reaction of the substrate molecule so that the end products (P) of the reaction are formed.
In summary: E + S —> ES –> E + P
E + S ⇄ ES ⇄ ES* ⇄ EP ⇄ E + P
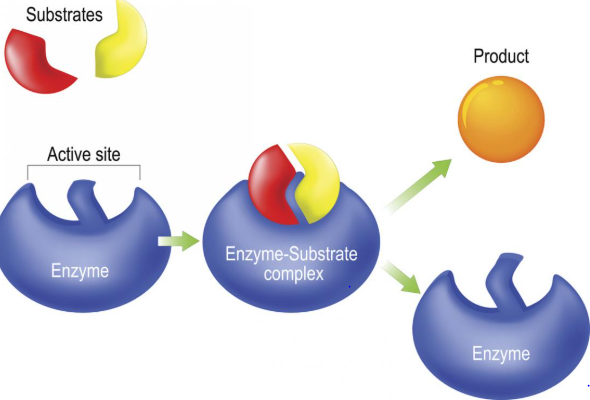
Fig 3: An enzyme molecule binds to the substrate. forming a complex called the enzyme substrate complex, on completion of the reaction the enzyme is released after the product is formed, the enzyme is not altered in the reaction and can even be recycled to break down into additional substrate molecules.
This, thereby makes a substrate more susceptible to certain reactions. Numerous changes such as this take place, the enzyme is released after the product is formed
5.4 Enzyme classification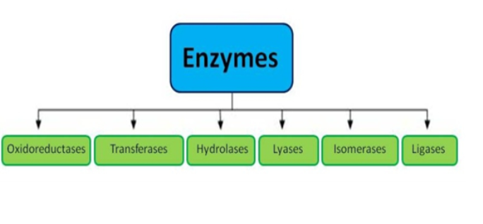
3). Hydrolases: these enzymes catalyse the hydrolysis or breakdown of the substrate.
E.g., Lysozyme, digestive enzymes, acid phosphatase.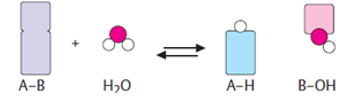
thereby facilitating the flow of the reaction.
Types | Biochemical Property |
Oxidoreductases | The enzyme Oxidoreductase catalyses the oxidation reaction in this reaction the electrons tend to travel from one form of a molecule to the other. |
Transferases | Functional groups such as donor and acceptor molecules, are transported with the help of enzyme Transferases enzymes. |
Hydrolases | Hydrolases are hydrolytic enzymes, which catalyse the hydrolysis reaction by adding water to cleave the bond and hydrolyse it. |
Lyases | These enzymes eliminate water, carbon dioxide or ammonia or create double bonds or add water, carbon dioxide or ammonia across double bonds. |
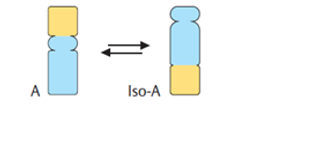
Isomerases | The Isomerases enzymes catalyse the structural shifts present in a molecule, thus causing the change in the shape of the molecule. |
Ligases | The Ligases enzymes are known to charge the catalysis of a ligation process. |
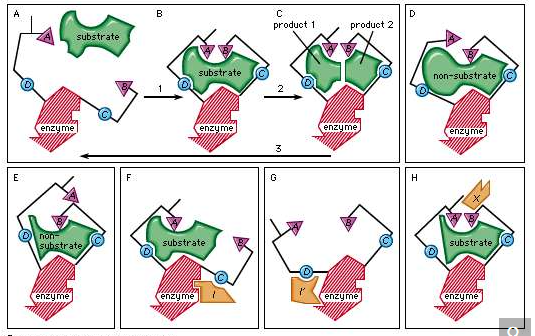
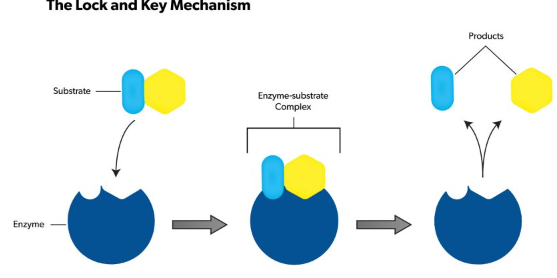
3. The lock-and-key model refers to the way similar to a lock and key mechanism, as each key is specific to particular lock, in this model also, a substrate binds to an enzyme's active site. They look the same like a key has to .be specific one for a lock, no reaction takes place if an incorrect substrate tries to bind to a different enzyme.
4. The active site present in an enzyme is a specific region that receives the substrate. It possesses a unique shape that complements that of the substrate, allowing for specificity to only one or two compounds. The substrate binds to the active site of the enzyme with a particular shape, and a reaction takes place that ultimately causes the release of the product formed.
5. Enzymes catalyse this reaction by easing chemical bond changes in the substrate by altering the distribution of electrons. Once the product has been released, the enzyme regenerates itself and is, ready for another reaction cycle.
6. The lock-and-key analogy sees this process as very specific process, further only a particular key can fit into the keyhole of the specific lock. If the key is in any way a little bigger or smaller, or entirely a different shape, then it does not fit into the specific keyhole, and subsequently the reaction cannot occur. The theory was first described by Emile Fischer (lock-and-key analogy) in 1894, and since then many other theories to were discovered to explain the mechanics of enzyme reactions.
2. Plotting Vi as a function of [S], the following is observed
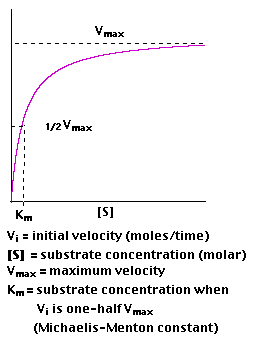
3. Km is roughly defined as the measure which is inverse to the strength or affinity of binding between the enzyme and its substrate. The lower the Km, the greater will be the affinity (so the lower the concentration of substrate needed to achieve a given rate of reaction).
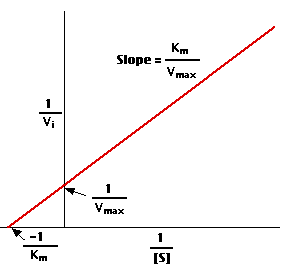
4. A "double-reciprocal" or Lineweaver-Burk plot is obtained by plotting the reciprocals of the same data points. Vmax and Km is determined precisely by this way which shows accuracy.
5. The Effects of Enzyme Inhibitors
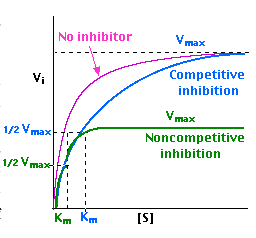
Enzymes can be inhibited in various ways
With non-competitive inhibition, enzyme molecules that have been bound by the inhibitor are not taken into consideration.
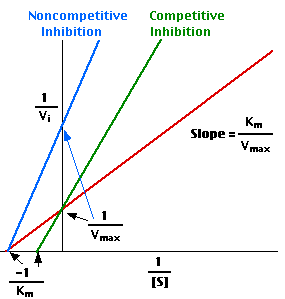
6. Enzymes are protein catalysts that, like all catalysts, speed up the rate of a chemical reaction without being themselves used up in the process.
7. They achieve their effect by temporarily binding to the substrate and, thereby, lower the activation energy needed to convert it to a product.
- competitive inhibitors are molecules that bind to the same site as the substrate — preventing the substrate from binding to the enzyme active site — but are not changed by the enzyme.
- non-competitive inhibitors are molecules that bind to some other site on the enzyme reducing the power of the catalysis.
4. The larger and smaller subunits split apart, when a ribosome finishes reading an mRNA molecule. Ribosomes are ribozymes, because the catalytic peptyl transferase activity that links amino acids together is performed by the ribosomal RNA.
5. The rough Endoplasmic reticulum Ribosomes present in the three-domain system namely the bacteria, Archaea and eukaryotes that have a common origin and a lot of similarities are often associated with the ribosomal RNA They differ in their structure, size, sequence, and the ratio of protein to RNA. The differences in structure of these cells can permit some antibiotics to kill bacteria by inhibiting their ribosomes, however human ribosomes remain unaffected.
6. However, in few cases one or more than one ribosome moves along a single mRNA at one time also known as polysome. which is found in all species, each "reading" its sequence and producing a corresponding protein molecule.
7. The mitochondrial ribosomes present in eukaryotic cells functions resemble many features of those in bacteria, reflecting the likely evolutionary origin of mitochondria.
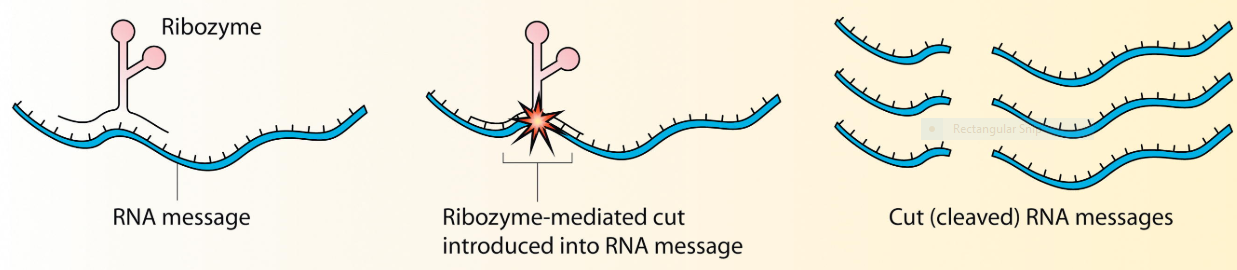
Fig 6: schematic showing ribosome cleavage of RNA
References: