BIO1
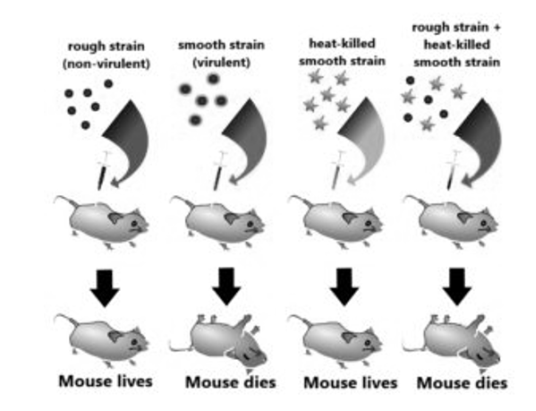
Module-6Information transfer Q1. Explain the discovery of Griffith?A1. While working with Streptococcus pneumoniae (the bacterium that causes pneumonia) in 1928, Frederick Griffith observed something nothing short of a miracle. He observed a transformation in this bacterium. When this bacterium is grown on a culture plate, after incubation some produce colonies that were shiny in nature (denoted as ‘S’) while some produce rough colonies (denoted as ‘R’).The S strain bacteria have a specific coat known as the polysaccharide coat which gives rise to smooth, shiny colonies. The R strain lacks this coat and hence, it gives rough colonies. Also, the S strain is virulent and causes pneumonia; while the R strain is non-virulent. He performed the following experiment with these strains and saw different observations.S strain → Inject into mice → Mice develop pneumonia and die. R strain → Inject into mice → Mice live. Heat-killed S strain → Inject into mice → Mice live. (Griffith found that heating kills the bacteria). Heat-killed S strain + R strain → Inject into mice → Mice die.
This is the experiment conducted by Griffith that determined the ‘Transforming principle that transformed from the S strain to R strain.Observations – Not only did the mice injected with the heat-killed S strain + R strain die, but Griffith most importantly recovered the live S strain bacteria from these dead mice.Conclusions – He concluded that this was because the R strain had somehow been ‘transformed’ by the heat -killed S strain. This he argued was due to the transfer of a ‘transforming principle ‘from the S strain to the R strain, which made the R strain virulent. Although significant, his observations did not identify the biochemical nature of the transforming principle. Q2. Illustrate the observations of Hershey and Chase?A2. Much earlier, scientists believed that the genetic material was protein. In 1952, Hershey & Chase were the ones to conclusively prove that DNA is the genetic material. They worked with bacteriophages – they are the viruses that infect bacteria. A bacteriophage attaches and transfers its genetic material into a bacterial cell, – This experiment conclusively showed that DNA is the genetic material transferred from virus to bacteria, and not protein.
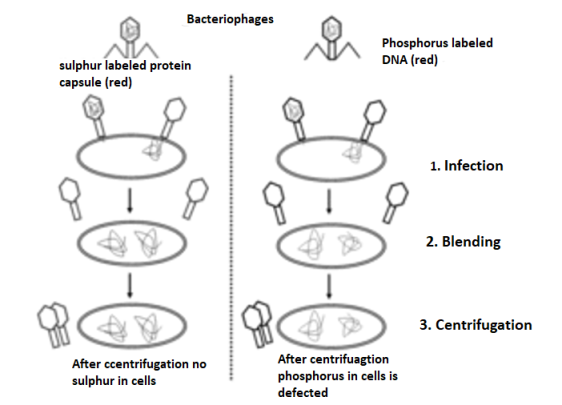
A phage is a small virus that infects bacteria. Its structure consists of a protein coat that holds the inside the coat genetic material. When a phage infects a bacterium, it pushes its genetic material into the bacterium, while its coat remains outside.
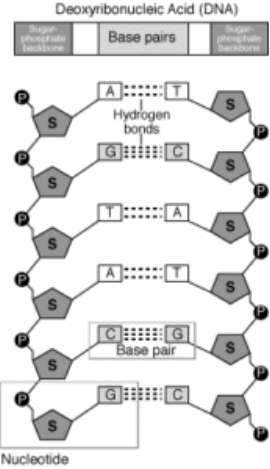
In a first experiment, T2 phages with radioactive 32P-labeled DNA were infected into bacteria. In a second experiment, T2 phages with radioactive 35S-labeled protein were into bacteria. In both experiments, bacteria were breakup from the phage coats by blending process followed by centrifugation. In the first experiment, most of the radioactivity was found in the infected bacteria, while in the second experiment most of the radioactivity was found in the phage coat. These experiments that were demonstrated proved that DNA is the genetic material of phage and that protein does not transmit genetic information.Q3.What is a Polynucleotide chain?A3. A nucleotide is made of the following components:Pentose sugar – A pentose sugar is a 5-carbon sugar compound. In case of DNA, this sugar is deoxyribose whereas, in RNA, it is ribose.Phosphate group Nitrogenous base – These can be of two types – Purines and Pyrimidines. Purines include Adenine and Guanine whereas pyrimidines include Cytosine and Thymine. In RNA, thymine is replaced by Uracil. Pentose sugar +Nitrogenous base + (via N-glycosidic linkage) = Nucleoside.Nucleoside + phosphate group (via phosphoester linkage) = Nucleotide.Nucleotide + Nucleotide (via 3′-5′ phosphodiester linkage) = Dinucleotide.Many nucleotides linked together = Polynucleotide.A polynucleotide has a free phosphate group at the 5′end position of the sugar and this is called the 5′ end. Similarly, the sugar also has a free 3′-OH group at the other end of the polynucleotide which is known as the 3′end. The backbone of a polynucleotide chain consists of phosphate groups and pentose sugars;however, the nitrogenous bases project out of this backbone.
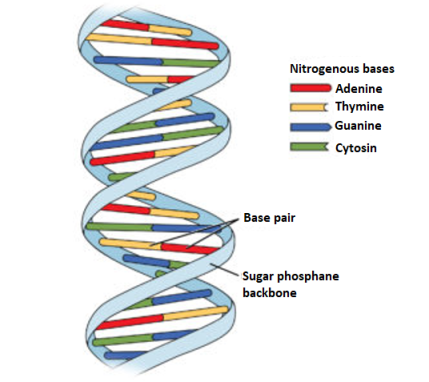
Polynucleotide of DNA and its componentsQ4.Explain the DNA double helix structure?A4. DNA is a long polymer and therefore, it makes it difficult to isolate from cells in an intact form. Therefore, it becomes difficult to study its structure. However, in 1953, James Watson and Francis introduced the ‘double helix’ model of the structure of DNA, from Maurice Wilkins and Rosalind Franklin based on their X-ray diffraction data. This model also reveals a unique property of polynucleotide chains – Base pairing. It refers to the hydrogen bonds that connect the nitrogen bases on opposite DNA strands. This pairing gives rise to complementary strands i.e. if the sequence of bases on one strand is known, the bases on the other strand can be predicted. further, if each DNA strand acts as a template for synthesis (parent) of a new strand, then the new double-stranded DNA (daughters) produced are identical to the parental DNA strand.Salient Features of DNA -HelixThe structure consists of two polynucleotide chains where the sugar and phosphate group form the backbone of the structure and the nitrogenous bases project inside the helix. The two polynucleotide chains have anti-parallel polarity i.e. if one strand has 5′ → 3′ polarity, on the other hand the other strand has 3′ → 5′ polarity. The bases on the opposite strands are connected through hydrogen bonds that form base pairs (bp). Adenine always binds to thymine with two hydrogen bonds from the opposite strand and the other way round. Guanine bonds with cytosine with three hydrogen bonds from the opposite strand and vice-versa. subsequently, a purine always pairs with a pyrimidine on the other strand, giving rise to a uniform distance between the two strands of the double helix. The two strands coil in a right-handed fashion. Each turn of the helix is 3.4nm (or 34 Angstrom units) that consist of 10 nucleotides. These nucleotides are placed at a distance of 0.34nm (or 3.4 Angstrom units). The helix is stable because of the base pairs that stack over one another and hydrogen bonds that hold the bases together.
The double helix structure of DNA, the bases on the opposite strands are connected through hydrogen bonds that form base pairs, the helix is stable because of the base pairs.Q5.Define a Nucleosome?A5. In Prokaryotes like E. coli, which do not have a well-defined nucleus. Here, the positively charged proteins and the negatively-charged DNA are held together in large loops in a structure called ‘nucleoid’. In Eukaryotes, however, the organization of DNA in the nucleus is much more complex and as seen below:The negatively-charged DNA is wrapped around a positively-charged (protein) histone octamer i.e.octamer aunit consisting of 8 histone molecules. This forms a structure called ‘Nucleosome ‘. Histones are positively-charged proteins that are rich in basic amino acids – arginines and lysines. A typical nucleosome has 200basepairs of DNA helix. Q6.Write a note on Genetic code?A6. The relationship between the sequence of amino acids in a polypeptide chain and nucleotide sequence of DNA or mRNA is called genetic code.Characteristics of Genetic codeTriplet code Start signal Stop signal Universal code Nonambiguous codons Related codons Commaless Polarity Degeneracy of code it presides over the transmission of genetic information to proteins, i.e., it makes up the protein synthesised by each cell by determines the sequence of amino acids in the polypeptide chain. Genetic information is coded in DNA by the of four bases: two purines (adenine and guanine) and two pyrimidines (thymine and cystosine). Each adjacent sequence of three bases (a codon) determines the insertion of a specific amino acid. In RNA, uracil replaces thymine the genetic information carried by the specific DNA molecules of the chromosomes; specifically, the system in particular combinations of three consecutive nucleotides in a DNA molecule control the insertion of one particular amino acid in equivalent places in a protein molecule. In plant and Animal kingdom the genetic code is almost universal throughout the prokaryotic. However, there are two known exceptions: In ciliated protozoans, the triplets AGA and AGG are read as termination signals instead of as L-arginine. This is also observed of in human mitochondrial code, where, in addition, uses AUA as a code for L-methionine (instead of L-isoleucine) and UGA for L-tryptophan (instead of a termination signal). The biochemical basis is that the set of DNA and RNA sequences that determine the amino acid sequences used in the synthesis of an organism's proteins, and nearly universal in all organisms. For protein synthesis a set of 64 codons corresponding to the 20 amino acids are used and as the signals for starting and stopping protein synthesis Q7. Explain complementation and recombination with an example?A7. When two mutants combine to restore the phenotype for a particular character the process is called Complementation. For example, when two mutant strains are mated it results in a wild type phenotype due to complementation. Thus, alleles which are wild type express its phenotype in offspring due to complementation effect. Moreover, the position of mutation determines importance of complementation. to determine if the gene is same or in a different gene is determined by a complementation test, in fact when mutations are present in different genes only then complementation is possible.
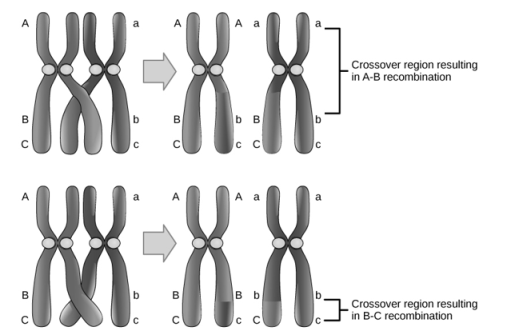
The experiment is an example of complementation process, where two mutants are mated that results is a wild Phenotype. The functionality of a particular pathway is determined by Complementation.Therefore, in various biochemical pathways the products are deduced by the phenomenon of complementation. Recombination is the process when the genetic material between two normal organisms are mixed to produce a recombinant organism or a mutant. The resulting mutant can be harmful or beneficial product. Moreover, the process of recombination is deliberately to done introduce positive characters to the new organism. In genetic recombination, the two parents contribute to form the mutant with altered genetic composition.
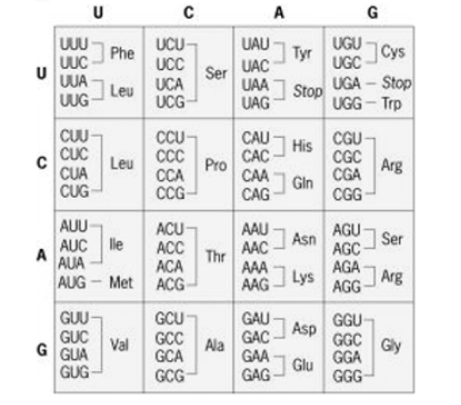
Shows recombination. that occurs when the genetic material between two normal organisms are mixed to produce a recombinant organism or a mutant Genetically modified organisms are produced by Recombination is a promising technique for the present era. The recombinant organism is produced through different techniques. Plasmids which act as microbial vector systems play a key role in recombination. In addition, bacteriophages are also used in genetic recombination. Furthermore, in genetic recombination physical mutagens such as radiation, chemicals are of great importance. In recombination, the two normal phenotype genes or organisms recombine to produce a genetically mutant organism. The reverse happens in Complementation where two genes or two organisms that are mutated complement each other and result in a genetically normal phenotype. During recombination, the mutant can either contain harmful characters or result in beneficial characters. Moreover, complementation is a more efficient technique in comparison to recombination. Thus, this summarizes the difference between complementation and recombination. Q8. Write a note on Degeneracy of Genetic code?A8. Degeneracy of Genetic Code:There are two methods by which the same amino acid is specified by two or more codons:1. The tRNAs accepting the same amino acid are different for different synonymous codons. “isoacceptortRNAs” is one such tRNA and they differ in anticodons. For example, one of the tRNAs carrying leucine is tRNA1leu with anticodon 3′ GAC5′, while the other is tRNA2leu with anticodon 3′ GAG5′.2. A single type of tRNA pairs with two or more synonymous codons. For example, tRNA. accepting the amino acid alanine in yeast (tRNAaIa) bears the anticodon 3′ CGI5′ that can pair with the codons 5′ GCU3′, 5′ GCC3 and 5′ GCA3′ on mRNA Crick in 1966 proposed the “wobble hypothesis” to explain the pairing of a single type anticodon with synonymous codons. The standard Genetic code
The standard Genetic code
According to the Wobble hypothesis, the base position at the 5′-end of anticodon is the “wobble position”. Two bases of anticodon from 3′-end are complementary to the two bases of the codon (in mRNA). The base at the wobble position can pair with different bases. For example, a single type of tRNAgly with the anticodon 3′ CCI5′ can pair with the codons 5’GGC3′ 5’GGU3′, and 5’GGA3′ specifying the amino acid glycine.Thus inosine (I) at the wobble position can pair, with C, U and A in the codon. Similarly, we can pair with A and G, while G at the wobble position can pair with C and/U.Q9. Write a note on universality of Genetic code?A9. The meaning of the universality of genetic code means that the same genetic code is utilized by all the organisms. For example, the lac+ gene producing the enzyme P-galactosidase, this enzyme is produced in human fibroblast tissue culture cells deficient of this enzyme is also seen in E. coli. When the haemoglobin mRNA molecules are injected into the Xenopus eggs, protein synthesis occurs and the α and β polypeptide chains are produced. However, variation in the genetic code has been observed in mitochondria where some of the codons are differently translated. The termination codon is UGA (termination codon in universal code) specifies tryptophan, similarly for AUA (for isoleucine in universal code) specifies methionine in mitochondria. Furthermore, the genetic code differs in the mitochondria of different organisms. For example, CUA is a codon for threonine in yeast mitochondria, while it specifies leucine in Drosophila and mammalian mitochondria.So, it may be concluded that the genetic code is not entirely universal. Q10. Define Chromatin?A10. Chromatin is composed of DNA and histones that are packaged into thin, stringy fibers. These chromatin fibers are not condensed but can exist in either a compact form (heterochromatin) or less compact form (euchromatin). Processes including DNA replication, transcription, and recombination occur in euchromatin. During cell division, chromatin condenses to form chromosomesChromatin is a mass of genetic material composed of DNA and proteins that condense to form chromosomes during eukaryotic cell division. Chromatin is located in the nucleus of our cells.The primary function of chromatin is to compress the DNA into a compact unit that will be less voluminous and can fit within the nucleus. Chromatin consists of complexes of small proteins known as histones and DNA
|
|
A phage is a small virus that infects bacteria. Its structure consists of a protein coat that holds the inside the coat genetic material. When a phage infects a bacterium, it pushes its genetic material into the bacterium, while its coat remains outside.
In a first experiment, T2 phages with radioactive 32P-labeled DNA were infected into bacteria. In a second experiment, T2 phages with radioactive 35S-labeled protein were into bacteria. In both experiments, bacteria were breakup from the phage coats by blending process followed by centrifugation. In the first experiment, most of the radioactivity was found in the infected bacteria, while in the second experiment most of the radioactivity was found in the phage coat. These experiments that were demonstrated proved that DNA is the genetic material of phage and that protein does not transmit genetic information.Q3.What is a Polynucleotide chain?A3. A nucleotide is made of the following components:Pentose sugar – A pentose sugar is a 5-carbon sugar compound. In case of DNA, this sugar is deoxyribose whereas, in RNA, it is ribose.
|
|
|
|
 The standard Genetic code
The standard Genetic code
|
0 matching results found